# ! pip install uniPairs statsmodels lifelines
from uniPairs import UniPairs, UniPairsTwoStage
import numpy as np
import pandas as pd
from sklearn.metrics import roc_auc_score
from lifelines.utils import concordance_index
from lifelines import CoxPHFitter
import statsmodels.api as sm
from IPython.display import display
# -----------------------------
# Synthetic Data Generation following Yu et al. Reluctant Interaction Modeling
# -----------------------------
def generate_synthetic_data(
n_train,
n_test,
p,
rho,
structure,
snr,
family,
censoring_rate=0.3,
seed=305,
):
rng = np.random.default_rng(seed)
if structure == "mixed":
T1 = [0,1,2,3,4,5]
T3 = [(0,4),(3,17),(9,10),(8,16),(0,12),(3,16)]
elif structure == "hierarchical":
T1 = [0,1,2,3,4,5]
T3 = [(0,2),(1,3),(2,3),(0,7),(1,7),(4,9)]
elif structure == "anti_hierarchical":
T1 = [0,1,2,3,4,5]
T3 = [(10,12),(11,13),(12,13),(10,17),(11,17),(14,19)]
elif structure == "interaction_only":
T1 = []
T3 = [(0,2),(1,3),(2,3),(0,7),(1,7),(4,9)]
elif structure == "main_only":
T1 = [0,1,2,3,4,5]
T3 = []
elif structure == "two_feature":
T1 = [0,1]
T3 = [(0,1)]
elif structure == "unit_test":
T1 = [0,1,2]
T3 = [(0,1),(1,2)]
else:
raise ValueError(f"Unknown structure '{structure}'")
max_idx = max(T1 + [i for pair in T3 for i in pair], default=0)
if p <= max_idx:
raise ValueError(f"Need p >= {max_idx} for structure '{structure}'")
idx = np.arange(p)
cov = rho ** np.abs(np.subtract.outer(idx, idx))
X = rng.multivariate_normal(
mean=np.zeros(p),
cov=cov,
size=n_train + n_test
)
X_true = np.hstack([X[:, j:j+1] for j in T1] +
[X[:, j:j+1] * X[:, k:k+1] for (j, k) in T3])
X_all_main_true_int = np.hstack(
[X] + [X[:, j:j+1] * X[:, k:k+1] for (j, k) in T3]
)
beta = np.zeros(p)
for j in T1:
beta[j] = 2.0
def interaction_signal(X_):
s = np.zeros(X_.shape[0])
for (j, k) in T3:
s += 3.0 * X_[:, j] * X_[:, k]
return s
mu_main = X @ beta
mu_int = interaction_signal(X)
if len(T1) > 0:
F = X[:, T1]
mu_proj = F @ np.linalg.pinv(F.T @ F) @ (F.T @ mu_int)
mu_main += mu_proj
mu_int -= mu_proj
var_main, var_int = np.var(mu_main), np.var(mu_int)
if var_main > 0 and var_int > 0:
mu_int *= np.sqrt(var_main / var_int)
mu_full = mu_main + mu_int
r2_main, r2_full = None, None
if family == "gaussian":
noise = rng.normal(
loc=0.0,
scale=np.sqrt(np.var(mu_full) / snr),
size=n_train + n_test
)
Y = mu_full + noise
var_Y = np.var(Y)
r2_main = 1 - np.mean((Y - mu_main) ** 2) / var_Y
r2_full = 1 - np.mean((Y - mu_full) ** 2) / var_Y
elif family == "binomial":
eta = mu_full / np.std(mu_full) * np.sqrt(snr)
p_true = 1 / (1 + np.exp(-eta))
Y = rng.binomial(1, p_true).astype(float)
elif family == "cox":
eta = mu_full / np.std(mu_full) * np.sqrt(snr)
lambda_ = np.exp(eta)
U = rng.uniform(size=n_train + n_test)
T = -np.log(U) / lambda_
C = rng.exponential(
scale=np.quantile(T, 1 - censoring_rate),
size=n_train + n_test
)
time = np.minimum(T, C)
status = (T <= C).astype(int)
Y = np.column_stack((time, status))
else:
raise ValueError(f"Unknown family '{family}'")
X_train, X_test = X[:n_train], X[n_train:]
Y_train, Y_test = Y[:n_train], Y[n_train:]
X_true_train, X_true_test = X_true[:n_train], X_true[n_train:]
X_all_main_true_int_train = X_all_main_true_int[:n_train]
X_all_main_true_int_test = X_all_main_true_int[n_train:]
return {
"X_train": X_train,
"Y_train": Y_train,
"X_test": X_test,
"Y_test": Y_test,
"R2_main": r2_main,
"R2_full": r2_full,
"X_true_train": X_true_train,
"X_true_test": X_true_test,
"X_all_main_true_int_train": X_all_main_true_int_train,
"X_all_main_true_int_test": X_all_main_true_int_test,
"true_active_main": [f"X{i}" for i in T1],
"true_active_int": [f"X{j}*X{k}" for (j, k) in T3],
"true_active_vars": [f"X{i}" for i in T1] +
[f"X{j}*X{k}" for (j, k) in T3],
"signal": " + ".join(
[f"2*X{i}" for i in T1] +
[f"3*X{j}*X{k}" for (j, k) in T3]
),
}
Gaussian¶
# -----------------------------
# Synthetic Gaussian Data
# -----------------------------
n_train=300
n_test=1000
p=400
rho=0.5
structure="mixed"
snr=1.
gaussian_synthetic_data = generate_synthetic_data(n_train=n_train, n_test=n_test, p=p, rho=rho, structure=structure, snr=snr, family="gaussian")
X_train = gaussian_synthetic_data['X_train']
Y_train = gaussian_synthetic_data['Y_train']
X_test = gaussian_synthetic_data['X_test']
Y_test = gaussian_synthetic_data['Y_test']
print(f"X_train.shape = {X_train.shape}, Y_train.shape = {Y_train.shape}")
print(f"X_test.shape = {X_test.shape}, Y_test.shape = {Y_test.shape}")
true_active_vars = gaussian_synthetic_data['true_active_vars']
print(f"True active vars: {true_active_vars}")
R2_main = gaussian_synthetic_data['R2_main']
R2_full = gaussian_synthetic_data['R2_full']
print(f"R2_main = {R2_main:.3f}, R2_full = {R2_full:.3f}")
X_train.shape = (300, 400), Y_train.shape = (300,)
X_test.shape = (1000, 400), Y_test.shape = (1000,)
True active vars: ['X0', 'X1', 'X2', 'X3', 'X4', 'X5', 'X0*X4', 'X3*X17', 'X9*X10', 'X8*X16', 'X0*X12', 'X3*X16']
R2_main = 0.246, R2_full = 0.511
# -----------------------------
# Fitting UniPairs-2stage
# -----------------------------
model = UniPairs(
two_stage=True,
hierarchy=None,
lmda_path_main_effects=None,
lmda_path_interactions=None,
n_folds_main_effects=10,
n_folds_interactions=10,
plot_cv_curve=True,
cv1se=False,
verbose=True,
interaction_candidates=None,
interaction_pairs=None
)
model.fit(X_train, Y_train)
pred_active_vars = model.get_active_variables()
formula = model.get_fitted_function()
alg_results = {
'UniPairsTwoStage/Test_R2': 1 - np.mean((Y_test - model.predict(X_test)) ** 2) / np.var(Y_test),
'UniPairsTwoStage/Train_R2': 1 - np.mean((Y_train - model.predict(X_train)) ** 2) / np.var(Y_train),
'UniPairsTwoStage/Coverage': len(set(pred_active_vars) & set(true_active_vars)) / len(true_active_vars),
'UniPairsTwoStage/Model_size': len(pred_active_vars),
'UniPairsTwoStage/FDP' : len(set(pred_active_vars) - set(true_active_vars)) / len(pred_active_vars) if len(pred_active_vars)>0 else 0,
'UniPairsTwoStage/Formula' : formula,
}
for k, v in alg_results.items():
print(f"{k}: {v}")
=== Starting UniPairs-2stage fit with 400 features ===
[Stage 1] Fitting main effects with UniLasso...
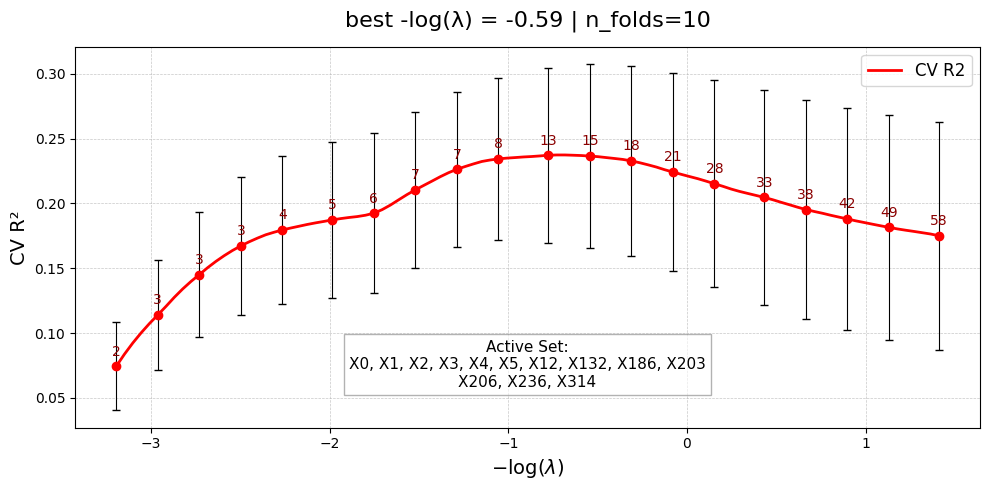
[Stage 1] Done. Active main effects: 13/400
Fitting triplet models ...
Fitting 79800 triplet models...
Progress: 7980/79800 triplets fitted...
Progress: 15960/79800 triplets fitted...
Progress: 23940/79800 triplets fitted...
Progress: 31920/79800 triplets fitted...
Progress: 39900/79800 triplets fitted...
Progress: 47880/79800 triplets fitted...
Progress: 55860/79800 triplets fitted...
Progress: 63840/79800 triplets fitted...
Progress: 71820/79800 triplets fitted...
Progress: 79800/79800 triplets fitted...
Triplet models complete. 0 unstable.
[Stage 2] Fitting interactions with Lasso...
Scanning interactions ...
Selected 1 interaction pairs. (largest log-gap rule)
Constructing interaction matrix with # pairs = 1
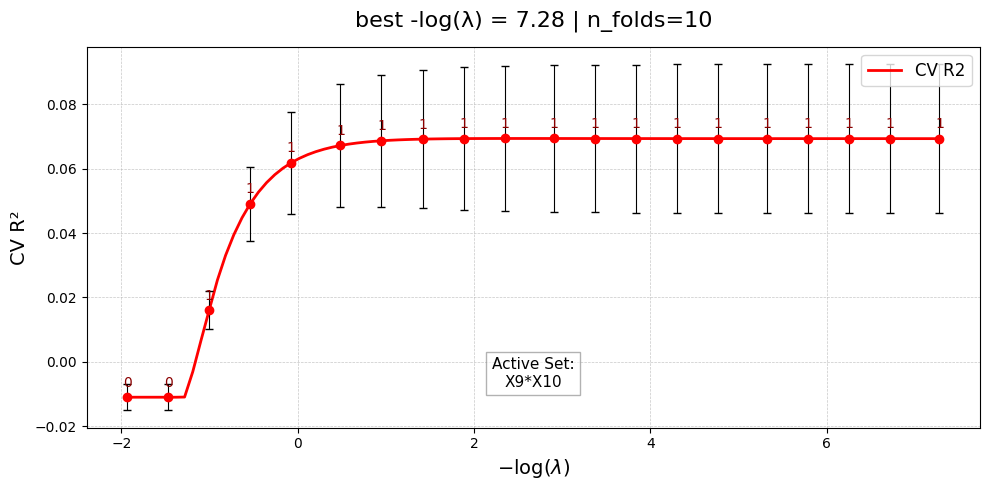
[Stage 2] Done. Active interactions: 1/1
=== UniPairs-2stage fit complete ===
Constructing interaction matrix with # pairs = 1
Constructing interaction matrix with # pairs = 1
UniPairsTwoStage/Test_R2: 0.2615194162959287
UniPairsTwoStage/Train_R2: 0.4317226168476088
UniPairsTwoStage/Coverage: 0.5833333333333334
UniPairsTwoStage/Model_size: 16
UniPairsTwoStage/FDP: 0.5625
UniPairsTwoStage/Formula: 1.066 + 1.294*X0 + 2.203*X1 + 2.673*X2 + 2.897*X3 + 0.866*X4 + 0.315*X5 + -0.042*X9 + 0.029*X10 + 0.462*X12 + 0.277*X132 + 0.514*X186 + 0.311*X203 + -0.229*X206 + -1.432*X236 + 1.198*X314 + -1.886 + 3.238*X9*X10
# -----------------------------
# Fitting UniPairs
# -----------------------------
model = UniPairs(
two_stage=False,
lmda_path=None,
n_folds=10,
plot_cv_curve=True,
cv1se=False,
verbose=True,
interaction_candidates=None,
interaction_pairs=None
)
model.fit(X_train, Y_train)
pred_active_vars = model.get_active_variables()
formula = model.get_fitted_function()
alg_results = {
'UniPairsOneStage/Test_R2': 1 - np.mean((Y_test - model.predict(X_test)) ** 2) / np.var(Y_test),
'UniPairsOneStage/Train_R2': 1 - np.mean((Y_train - model.predict(X_train)) ** 2) / np.var(Y_train),
'UniPairsOneStage/Coverage': len(set(pred_active_vars) & set(true_active_vars)) / len(true_active_vars),
'UniPairsOneStage/Model_size': len(pred_active_vars),
'UniPairsOneStage/FDP' : len(set(pred_active_vars) - set(true_active_vars)) / len(pred_active_vars) if len(pred_active_vars)>0 else 0,
'UniPairsOneStage/Formula' : formula,
}
for k, v in alg_results.items():
print(f"{k}: {v}")
=== Starting UniPairs fit with 400 features ===
Fitting triplet models ...
Fitting 79800 triplet models...
Progress: 7980/79800 triplets fitted...
Progress: 15960/79800 triplets fitted...
Progress: 23940/79800 triplets fitted...
Progress: 31920/79800 triplets fitted...
Progress: 39900/79800 triplets fitted...
Progress: 47880/79800 triplets fitted...
Progress: 55860/79800 triplets fitted...
Progress: 63840/79800 triplets fitted...
Progress: 71820/79800 triplets fitted...
Progress: 79800/79800 triplets fitted...
Triplet models complete. 0 unstable.
Scanning interactions ...
Selected 1 interaction pairs. (largest log-gap rule)


Constructing interaction matrix with # pairs = 1
Cross-validating UniLasso ...
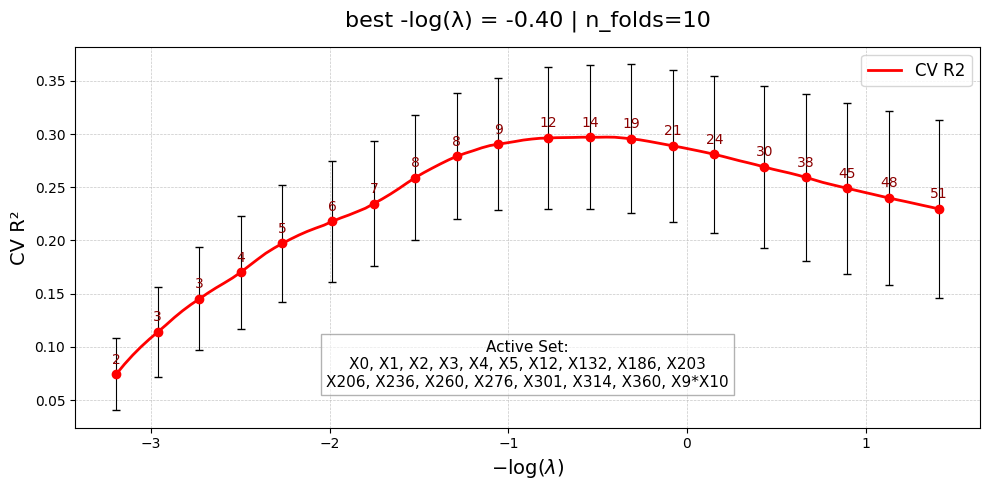
=== UniPairs fit complete ===
Constructing interaction matrix with # pairs = 1
Constructing interaction matrix with # pairs = 1
UniPairsOneStage/Test_R2: 0.25988452930239936
UniPairsOneStage/Train_R2: 0.44218385131195737
UniPairsOneStage/Coverage: 0.5833333333333334
UniPairsOneStage/Model_size: 20
UniPairsOneStage/FDP: 0.65
UniPairsOneStage/Formula: -0.292 + 1.380*X0 + 2.058*X1 + 2.609*X2 + 2.800*X3 + 1.141*X4 + 0.317*X5 + -0.034*X9 + 0.024*X10 + 0.720*X12 + 0.065*X132 + 0.557*X186 + 0.383*X203 + -0.195*X206 + -1.450*X236 + 0.120*X260 + -0.116*X276 + 0.065*X301 + 1.345*X314 + -0.154*X360 + 2.666*X9*X10
Cox¶
# -----------------------------
# Synthetic Cox Data
# -----------------------------
n_train=500
n_test=1000
p=20
rho=0.5
structure="mixed"
snr=1.
cox_synthetic_data = generate_synthetic_data(n_train=n_train, n_test=n_test, p=p, rho=rho, structure=structure, snr=snr, family="cox")
X_train = cox_synthetic_data['X_train']
Y_train = cox_synthetic_data['Y_train']
X_test = cox_synthetic_data['X_test']
Y_test = cox_synthetic_data['Y_test']
X_all_main_true_int_train = cox_synthetic_data['X_all_main_true_int_train']
X_all_main_true_int_test = cox_synthetic_data['X_all_main_true_int_test']
X_true_train = cox_synthetic_data['X_true_train']
X_true_test = cox_synthetic_data['X_true_test']
print(f"X_train.shape = {X_train.shape}, Y_train.shape = {Y_train.shape}")
print(f"X_test.shape = {X_test.shape}, Y_test.shape = {Y_test.shape}")
true_active_vars = cox_synthetic_data['true_active_vars']
true_active_main = cox_synthetic_data['true_active_main']
true_active_int = cox_synthetic_data['true_active_int']
print(f"True active vars: {true_active_vars}")
X_train.shape = (500, 20), Y_train.shape = (500, 2)
X_test.shape = (1000, 20), Y_test.shape = (1000, 2)
True active vars: ['X0', 'X1', 'X2', 'X3', 'X4', 'X5', 'X0*X4', 'X3*X17', 'X9*X10', 'X8*X16', 'X0*X12', 'X3*X16']
# -----------------------------
# Sanity check before fitting UniPairs:
# Fit a full Cox PH model using all main effects and the true interaction terms.
# If all true variables have extremely small p-values, the synthetic signal is too strong and the interaction-detection problem becomes trivial.
# -----------------------------
cph = CoxPHFitter()
cph_df_train = pd.DataFrame(np.hstack([X_all_main_true_int_train, Y_train]), columns=[f"X{i}" for i in range(p)]+true_active_int+['time', 'status'])
cph.fit(cph_df_train, duration_col='time', event_col='status')
display(cph.summary)
cph_df_test = pd.DataFrame(X_all_main_true_int_test, columns=[f"X{i}" for i in range(p)]+true_active_int)
print(f"Concordance index: {concordance_index(Y_test[:, 0], -cph.predict_partial_hazard(cph_df_test), Y_test[:, 1])}")
| coef | exp(coef) | se(coef) | coef lower 95% | coef upper 95% | exp(coef) lower 95% | exp(coef) upper 95% | cmp to | z | p | -log2(p) | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| covariate | |||||||||||
| X0 | 0.178232 | 1.195103 | 0.075624 | 0.030013 | 0.326452 | 1.030467 | 1.386042 | 0.0 | 2.356831 | 1.843164e-02 | 5.761671 |
| X1 | 0.244707 | 1.277247 | 0.081196 | 0.085566 | 0.403847 | 1.089334 | 1.497575 | 0.0 | 3.013790 | 2.580063e-03 | 8.598378 |
| X2 | 0.196804 | 1.217505 | 0.088843 | 0.022675 | 0.370932 | 1.022934 | 1.449085 | 0.0 | 2.215196 | 2.674665e-02 | 5.224498 |
| X3 | 0.281318 | 1.324875 | 0.089667 | 0.105575 | 0.457061 | 1.111349 | 1.579425 | 0.0 | 3.137379 | 1.704656e-03 | 9.196304 |
| X4 | 0.139807 | 1.150052 | 0.080434 | -0.017840 | 0.297454 | 0.982318 | 1.346426 | 0.0 | 1.738166 | 8.218159e-02 | 3.605041 |
| X5 | 0.158690 | 1.171974 | 0.085548 | -0.008981 | 0.326361 | 0.991059 | 1.385915 | 0.0 | 1.854979 | 6.359923e-02 | 3.974847 |
| X6 | 0.187387 | 1.206094 | 0.087358 | 0.016167 | 0.358606 | 1.016299 | 1.431333 | 0.0 | 2.145033 | 3.195022e-02 | 4.968030 |
| X7 | -0.096974 | 0.907580 | 0.084245 | -0.262090 | 0.068143 | 0.769442 | 1.070518 | 0.0 | -1.151096 | 2.496927e-01 | 2.001774 |
| X8 | -0.050891 | 0.950382 | 0.087658 | -0.222698 | 0.120916 | 0.800356 | 1.128530 | 0.0 | -0.580566 | 5.615333e-01 | 0.832557 |
| X9 | 0.031936 | 1.032451 | 0.084904 | -0.134474 | 0.198345 | 0.874175 | 1.219383 | 0.0 | 0.376135 | 7.068168e-01 | 0.500592 |
| X10 | 0.033196 | 1.033754 | 0.085528 | -0.134436 | 0.200829 | 0.874209 | 1.222415 | 0.0 | 0.388134 | 6.979169e-01 | 0.518873 |
| X11 | -0.007492 | 0.992536 | 0.081117 | -0.166477 | 0.151494 | 0.846642 | 1.163571 | 0.0 | -0.092359 | 9.264127e-01 | 0.110273 |
| X12 | -0.110949 | 0.894985 | 0.080551 | -0.268825 | 0.046928 | 0.764277 | 1.048047 | 0.0 | -1.377374 | 1.683966e-01 | 2.570065 |
| X13 | -0.024206 | 0.976084 | 0.081663 | -0.184263 | 0.135851 | 0.831717 | 1.145511 | 0.0 | -0.296416 | 7.669126e-01 | 0.382866 |
| X14 | 0.109636 | 1.115872 | 0.090914 | -0.068551 | 0.287824 | 0.933746 | 1.333522 | 0.0 | 1.205940 | 2.278406e-01 | 2.133903 |
| X15 | -0.054161 | 0.947279 | 0.085125 | -0.221004 | 0.112681 | 0.801713 | 1.119275 | 0.0 | -0.636252 | 5.246119e-01 | 0.930678 |
| X16 | -0.047890 | 0.953239 | 0.086477 | -0.217381 | 0.121602 | 0.804623 | 1.129305 | 0.0 | -0.553784 | 5.797267e-01 | 0.786555 |
| X17 | -0.083190 | 0.920176 | 0.090305 | -0.260184 | 0.093804 | 0.770910 | 1.098344 | 0.0 | -0.921216 | 3.569378e-01 | 1.486255 |
| X18 | -0.004735 | 0.995276 | 0.093032 | -0.187074 | 0.177604 | 0.829383 | 1.194352 | 0.0 | -0.050896 | 9.594083e-01 | 0.059783 |
| X19 | -0.043288 | 0.957636 | 0.074194 | -0.188706 | 0.102131 | 0.828030 | 1.107528 | 0.0 | -0.583437 | 5.595990e-01 | 0.837535 |
| X0*X4 | 0.270650 | 1.310816 | 0.065722 | 0.141836 | 0.399463 | 1.152388 | 1.491025 | 0.0 | 4.118073 | 3.820533e-05 | 14.675866 |
| X3*X17 | 0.380933 | 1.463649 | 0.075699 | 0.232564 | 0.529301 | 1.261832 | 1.697745 | 0.0 | 5.032168 | 4.849628e-07 | 20.975622 |
| X9*X10 | 0.300783 | 1.350917 | 0.057188 | 0.188698 | 0.412869 | 1.207676 | 1.511147 | 0.0 | 5.259599 | 1.443698e-07 | 22.723728 |
| X8*X16 | 0.261423 | 1.298777 | 0.069424 | 0.125354 | 0.397492 | 1.133550 | 1.488088 | 0.0 | 3.765592 | 1.661550e-04 | 12.555183 |
| X0*X12 | 0.380720 | 1.463338 | 0.072951 | 0.237739 | 0.523701 | 1.268379 | 1.688265 | 0.0 | 5.218867 | 1.800210e-07 | 22.405331 |
| X3*X16 | 0.303419 | 1.354481 | 0.082484 | 0.141754 | 0.465083 | 1.152293 | 1.592147 | 0.0 | 3.678535 | 2.345772e-04 | 12.057649 |
Concordance index: 0.7315450981420536
# -----------------------------
# Fitting UniPairs-2stage
# -----------------------------
model = UniPairs(
two_stage=True,
hierarchy=None,
lmda_path_main_effects=None,
lmda_path_interactions=None,
n_folds_main_effects=10,
n_folds_interactions=10,
plot_cv_curve=True,
cv1se=False,
verbose=True,
interaction_candidates=None,
interaction_pairs=None,
family_spec={'family':'cox'}
)
model.fit(X_train, Y_train)
pred_active_vars = model.get_active_variables()
formula = model.get_fitted_function()
alg_results = {
'UniPairsTwoStage/Test_c_index' : concordance_index(Y_test[:, 0], -model.predict(X_test), Y_test[:, 1]),
'UniPairsTwoStage/Train_c_index' : concordance_index(Y_train[:, 0], -model.predict(X_train), Y_train[:, 1]),
'UniPairsTwoStage/Coverage': len(set(pred_active_vars) & set(true_active_vars)) / len(true_active_vars),
'UniPairsTwoStage/Model_size': len(pred_active_vars),
'UniPairsTwoStage/FDP' : len(set(pred_active_vars) - set(true_active_vars)) / len(pred_active_vars) if len(pred_active_vars)>0 else 0,
'UniPairsTwoStage/Formula' : formula,
}
for k, v in alg_results.items():
print(f"{k}: {v}")
/Users/aymen20/Desktop/Research/repo/pkgs/uniPairs/.venv/lib/python3.9/site-packages/adelie/matrix.py:648: UserWarning: Detected matrix to be C-contiguous. Performance may improve with F-contiguous matrix.
warnings.warn(
100%|██████████| 100/100 [00:00:00<00:00:00, 5072.46it/s] [dev:14.9%]
100%|██████████| 103/103 [00:00:00<00:00:00, 4589.75it/s] [dev:15.9%]
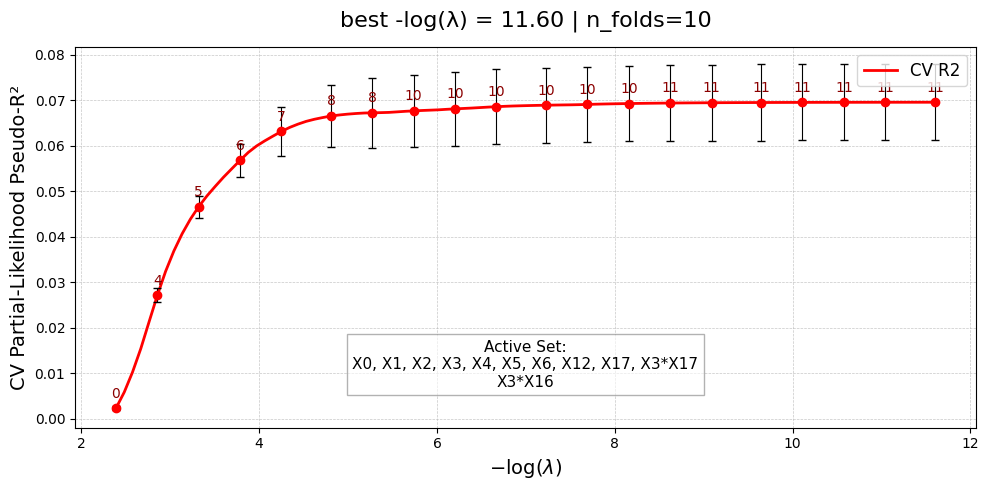
=== Starting UniPairs-2stage fit with 20 features ===
[Stage 1] Fitting main effects with UniLasso...
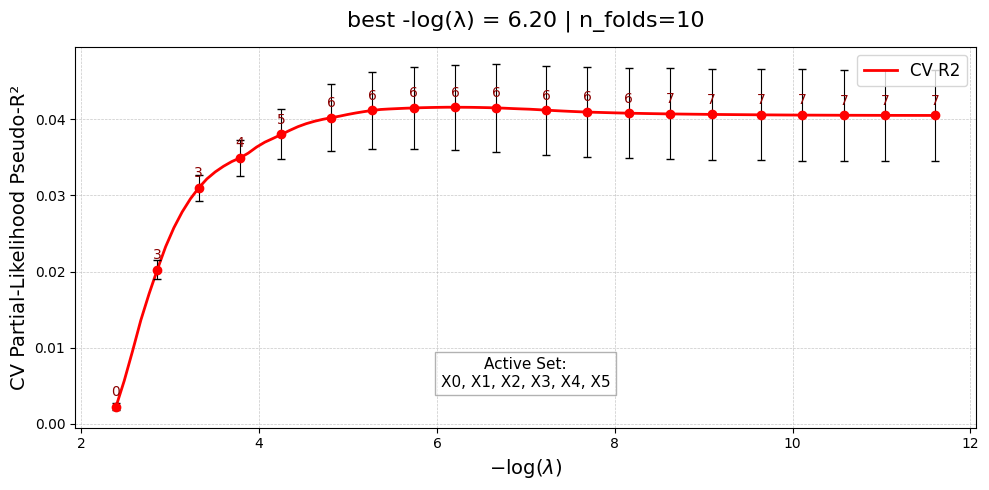
[Stage 1] Done. Active main effects: 6/20
Fitting triplet models ...
Fitting 190 triplet models...
Progress: 19/190 triplets fitted...
Progress: 38/190 triplets fitted...
Progress: 57/190 triplets fitted...
Progress: 76/190 triplets fitted...
Progress: 95/190 triplets fitted...
Progress: 114/190 triplets fitted...
Progress: 133/190 triplets fitted...
Progress: 152/190 triplets fitted...
Progress: 171/190 triplets fitted...
Progress: 190/190 triplets fitted...
Triplet models complete. 0 unstable.
[Stage 2] Fitting interactions with Lasso...
Scanning interactions ...
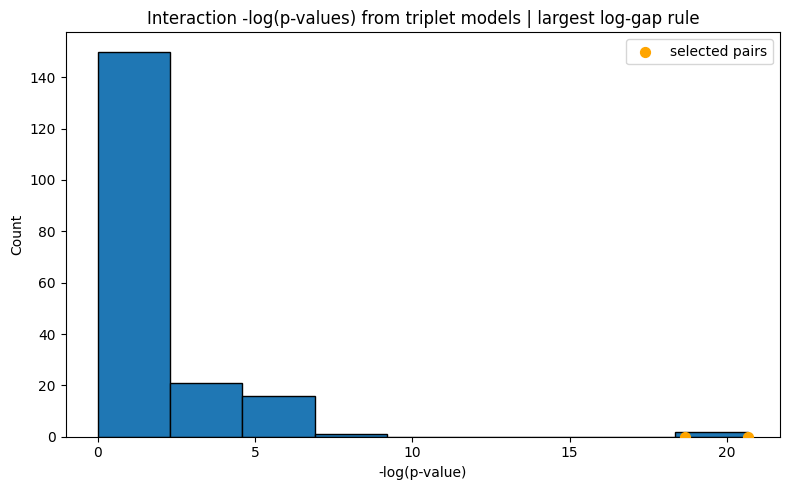
Selected 2 interaction pairs. (largest log-gap rule)

/Users/aymen20/Desktop/Research/repo/pkgs/uniPairs/.venv/lib/python3.9/site-packages/adelie/matrix.py:648: UserWarning: Detected matrix to be C-contiguous. Performance may improve with F-contiguous matrix.
warnings.warn(
100%|██████████| 100/100 [00:00:00<00:00:00, 20287.92it/s] [dev:1.1%]
100%|██████████| 102/102 [00:00:00<00:00:00, 19350.55it/s] [dev:2.8%]
Constructing interaction matrix with # pairs = 2
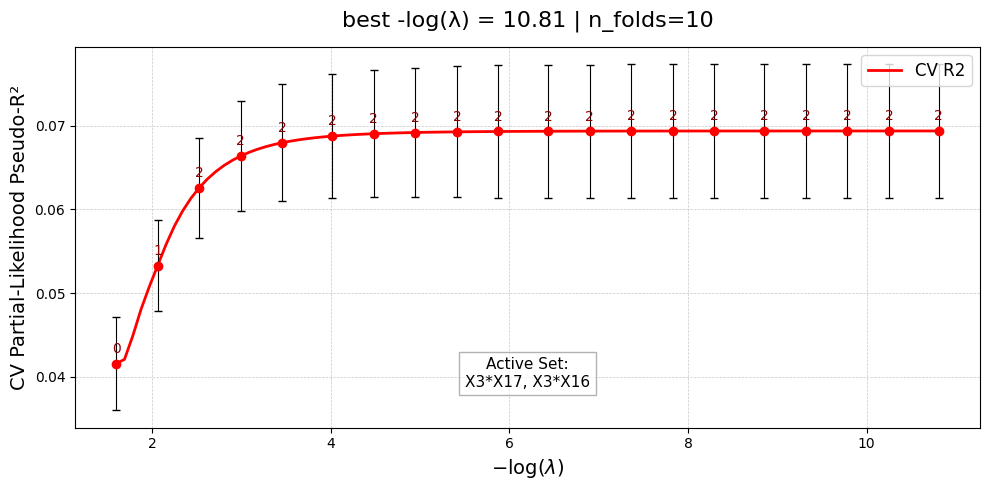
[Stage 2] Done. Active interactions: 2/2
=== UniPairs-2stage fit complete ===
Constructing interaction matrix with # pairs = 2
Constructing interaction matrix with # pairs = 2
UniPairsTwoStage/Test_c_index: 0.6711651172853266
UniPairsTwoStage/Train_c_index: 0.7011771147002464
UniPairsTwoStage/Coverage: 0.6666666666666666
UniPairsTwoStage/Model_size: 10
UniPairsTwoStage/FDP: 0.2
UniPairsTwoStage/Formula: -0.004 + 0.123*X0 + 0.235*X1 + 0.137*X2 + 0.232*X3 + 0.038*X4 + 0.167*X5 + -0.007*X16 + -0.012*X17 + -0.000 + 0.288*X3*X17 + 0.176*X3*X16
# -----------------------------
# Fitting UniPairs
# -----------------------------
model = UniPairs(
two_stage=False,
lmda_path=None,
n_folds=10,
plot_cv_curve=True,
cv1se=False,
verbose=True,
interaction_candidates=None,
interaction_pairs=None,
family_spec={'family':'cox'}
)
model.fit(X_train, Y_train)
pred_active_vars = model.get_active_variables()
formula = model.get_fitted_function()
alg_results = {
'UniPairsOneStage/Test_c_index' : concordance_index(Y_test[:, 0], -model.predict(X_test), Y_test[:, 1]),
'UniPairsOneStage/Train_c_index' : concordance_index(Y_train[:, 0], -model.predict(X_train), Y_train[:, 1,]),
'UniPairsOneStage/Coverage': len(set(pred_active_vars) & set(true_active_vars)) / len(true_active_vars),
'UniPairsOneStage/Model_size': len(pred_active_vars),
'UniPairsOneStage/FDP' : len(set(pred_active_vars) - set(true_active_vars)) / len(pred_active_vars) if len(pred_active_vars)>0 else 0,
'UniPairsOneStage/Formula' : formula,
}
for k, v in alg_results.items():
print(f"{k}: {v}")
=== Starting UniPairs fit with 20 features ===
Fitting triplet models ...
Fitting 190 triplet models...
Progress: 19/190 triplets fitted...
Progress: 38/190 triplets fitted...
Progress: 57/190 triplets fitted...
Progress: 76/190 triplets fitted...
Progress: 95/190 triplets fitted...
Progress: 114/190 triplets fitted...
Progress: 133/190 triplets fitted...
Progress: 152/190 triplets fitted...
Progress: 171/190 triplets fitted...
Progress: 190/190 triplets fitted...
Triplet models complete. 0 unstable.
Scanning interactions ...
Selected 2 interaction pairs. (largest log-gap rule)


/Users/aymen20/Desktop/Research/repo/pkgs/uniPairs/.venv/lib/python3.9/site-packages/adelie/matrix.py:648: UserWarning: Detected matrix to be C-contiguous. Performance may improve with F-contiguous matrix.
warnings.warn(
100%|██████████| 100/100 [00:00:00<00:00:00, 4039.85it/s] [dev:19.3%]
100%|██████████| 103/103 [00:00:00<00:00:00, 4620.60it/s] [dev:14.0%]
Constructing interaction matrix with # pairs = 2
Cross-validating UniLasso ...
=== UniPairs fit complete ===
Constructing interaction matrix with # pairs = 2
Constructing interaction matrix with # pairs = 2
UniPairsOneStage/Test_c_index: 0.6692409561147022
UniPairsOneStage/Train_c_index: 0.701875171092253
UniPairsOneStage/Coverage: 0.6666666666666666
UniPairsOneStage/Model_size: 12
UniPairsOneStage/FDP: 0.3333333333333333
UniPairsOneStage/Formula: -0.003 + 0.122*X0 + 0.242*X1 + 0.123*X2 + 0.223*X3 + 0.106*X4 + 0.167*X5 + 0.069*X6 + -0.007*X12 + -0.009*X16 + -0.109*X17 + 0.294*X3*X17 + 0.216*X3*X16
Binomial¶
# -----------------------------
# Synthetic Logistic Data
# -----------------------------
n_train=500
n_test=1000
p=20
rho=0.5
structure="mixed"
snr=1.
binomial_synthetic_data = generate_synthetic_data(n_train=n_train, n_test=n_test, p=p, rho=rho, structure=structure, snr=snr, family="binomial")
X_train = binomial_synthetic_data['X_train']
Y_train = binomial_synthetic_data['Y_train']
X_test = binomial_synthetic_data['X_test']
Y_test = binomial_synthetic_data['Y_test']
print(f"X_train.shape = {X_train.shape}, Y_train.shape = {Y_train.shape}")
print(f"X_test.shape = {X_test.shape}, Y_test.shape = {Y_test.shape}")
true_active_vars = binomial_synthetic_data['true_active_vars']
print(f"True active vars: {true_active_vars}")
X_train.shape = (500, 20), Y_train.shape = (500,)
X_test.shape = (1000, 20), Y_test.shape = (1000,)
True active vars: ['X0', 'X1', 'X2', 'X3', 'X4', 'X5', 'X0*X4', 'X3*X17', 'X9*X10', 'X8*X16', 'X0*X12', 'X3*X16']
# ------------------------------------------------------------
# Sanity check before fitting UniPairs :
# Fit a logistic regression model using all main effects and the true interaction terms.
# If all true variables have extremely small p-values, then the
# synthetic signal is too strong and the interaction-detection task becomes trivial.
# ------------------------------------------------------------
X_all_main_true_int_train = binomial_synthetic_data['X_all_main_true_int_train']
X_all_main_true_int_test = binomial_synthetic_data['X_all_main_true_int_test']
X_true_train = binomial_synthetic_data['X_true_train']
X_true_test = binomial_synthetic_data['X_true_test']
true_active_main = binomial_synthetic_data['true_active_main']
true_active_int = binomial_synthetic_data['true_active_int']
binom_df_train = pd.DataFrame(np.hstack([X_all_main_true_int_train]), columns=[f"X{i}" for i in range(p)]+true_active_int)
model = sm.GLM(Y_train, sm.add_constant(binom_df_train), family=sm.families.Binomial())
res = model.fit()
print(res.summary())
binom_df_test = pd.DataFrame(np.hstack([X_all_main_true_int_test]), columns=[f"X{i}" for i in range(p)]+true_active_int)
res.predict(sm.add_constant(binom_df_test))
print(f"ROC AUC: {roc_auc_score(Y_test,res.predict(sm.add_constant(binom_df_test)))}")
Generalized Linear Model Regression Results
==============================================================================
Dep. Variable: y No. Observations: 500
Model: GLM Df Residuals: 473
Model Family: Binomial Df Model: 26
Link Function: Logit Scale: 1.0000
Method: IRLS Log-Likelihood: -278.28
Date: Sun, 14 Dec 2025 Deviance: 556.57
Time: 16:06:03 Pearson chi2: 498.
No. Iterations: 5 Pseudo R-squ. (CS): 0.2382
Covariance Type: nonrobust
==============================================================================
coef std err z P>|z| [0.025 0.975]
------------------------------------------------------------------------------
const -0.2116 0.115 -1.838 0.066 -0.437 0.014
X0 0.2148 0.125 1.714 0.087 -0.031 0.461
X1 0.3265 0.137 2.378 0.017 0.057 0.596
X2 0.3087 0.140 2.200 0.028 0.034 0.584
X3 0.1560 0.141 1.104 0.270 -0.121 0.433
X4 0.3169 0.144 2.196 0.028 0.034 0.600
X5 0.0676 0.134 0.503 0.615 -0.196 0.331
X6 0.1163 0.150 0.776 0.438 -0.177 0.410
X7 -0.2673 0.137 -1.945 0.052 -0.537 0.002
X8 -0.1288 0.141 -0.913 0.361 -0.405 0.148
X9 0.1549 0.143 1.084 0.278 -0.125 0.435
X10 -0.3055 0.138 -2.208 0.027 -0.577 -0.034
X11 0.1837 0.137 1.341 0.180 -0.085 0.452
X12 0.0849 0.134 0.634 0.526 -0.178 0.347
X13 -0.0251 0.134 -0.187 0.852 -0.288 0.238
X14 -0.3255 0.150 -2.175 0.030 -0.619 -0.032
X15 0.1044 0.143 0.730 0.465 -0.176 0.385
X16 0.0327 0.137 0.239 0.811 -0.235 0.301
X17 0.0891 0.137 0.652 0.514 -0.179 0.357
X18 0.0620 0.144 0.432 0.666 -0.220 0.344
X19 -0.1013 0.121 -0.836 0.403 -0.339 0.136
X0*X4 0.3805 0.122 3.127 0.002 0.142 0.619
X3*X17 0.3159 0.122 2.584 0.010 0.076 0.555
X9*X10 0.2952 0.107 2.770 0.006 0.086 0.504
X8*X16 0.1060 0.121 0.875 0.381 -0.131 0.343
X0*X12 0.4030 0.119 3.387 0.001 0.170 0.636
X3*X16 0.3447 0.128 2.694 0.007 0.094 0.595
==============================================================================
ROC AUC: 0.7147604563961442
# -----------------------------
# Fitting UniPairs-2stage
# -----------------------------
model = UniPairs(
two_stage=True,
hierarchy=None,
lmda_path_main_effects=None,
lmda_path_interactions=None,
n_folds_main_effects=10,
n_folds_interactions=10,
plot_cv_curve=True,
cv1se=False,
verbose=True,
interaction_candidates=None,
interaction_pairs=None,
family_spec={'family':'binomial'}
)
model.fit(X_train, Y_train)
pred_active_vars = model.get_active_variables()
formula = model.get_fitted_function()
alg_results = {
'UniPairsTwoStage/Test_ROC_AUC': roc_auc_score(Y_test, model.predict(X_test, response_scale=True)),
'UniPairsTwoStage/Train_ROC_AUC': roc_auc_score(Y_train, model.predict(X_train, response_scale=True)),
'UniPairsTwoStage/Coverage': len(set(pred_active_vars) & set(true_active_vars)) / len(true_active_vars),
'UniPairsTwoStage/Model_size': len(pred_active_vars),
'UniPairsTwoStage/FDP' : len(set(pred_active_vars) - set(true_active_vars)) / len(pred_active_vars) if len(pred_active_vars)>0 else 0,
'UniPairsTwoStage/Formula' : formula,
}
for k, v in alg_results.items():
print(f"{k}: {v}")
/Users/aymen20/Desktop/Research/repo/pkgs/uniPairs/.venv/lib/python3.9/site-packages/adelie/matrix.py:648: UserWarning: Detected matrix to be C-contiguous. Performance may improve with F-contiguous matrix.
warnings.warn(
100%|██████████| 102/102 [00:00:00<00:00:00, 6296.38it/s] [dev:55.9%]
100%|██████████| 100/100 [00:00:00<00:00:00, 8264.98it/s] [dev:50.9%]
=== Starting UniPairs-2stage fit with 20 features ===
[Stage 1] Fitting main effects with UniLasso...
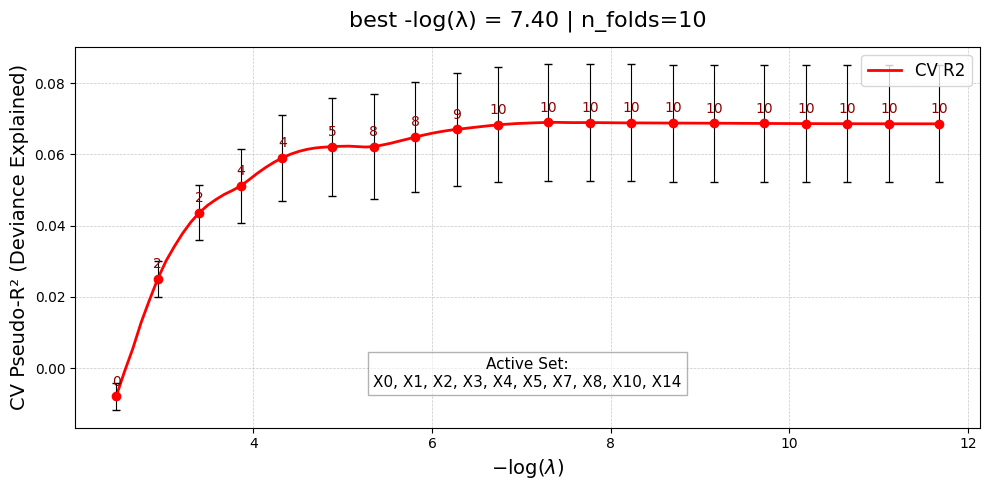
[Stage 1] Done. Active main effects: 10/20
Fitting triplet models ...
Fitting 190 triplet models...
Progress: 19/190 triplets fitted...
Progress: 38/190 triplets fitted...
Progress: 57/190 triplets fitted...
Progress: 76/190 triplets fitted...
Progress: 95/190 triplets fitted...
Progress: 114/190 triplets fitted...
Progress: 133/190 triplets fitted...
Progress: 152/190 triplets fitted...
Progress: 171/190 triplets fitted...
Progress: 190/190 triplets fitted...
Triplet models complete. 0 unstable.
[Stage 2] Fitting interactions with Lasso...
Scanning interactions ...
Selected 3 interaction pairs. (largest log-gap rule)
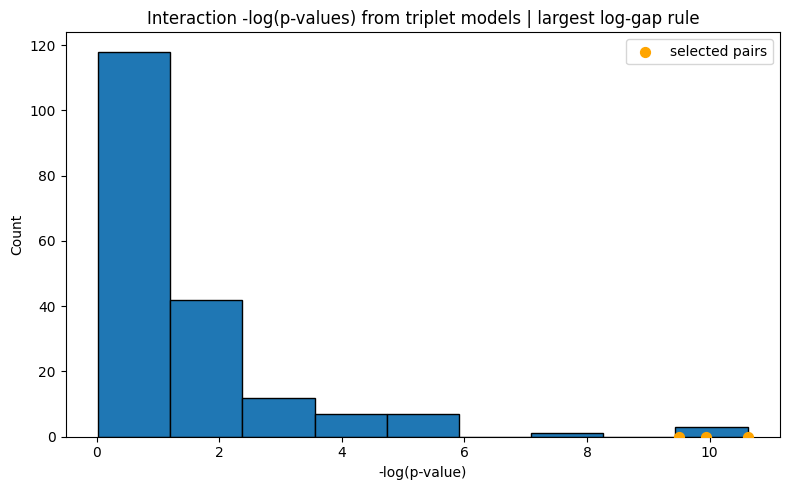
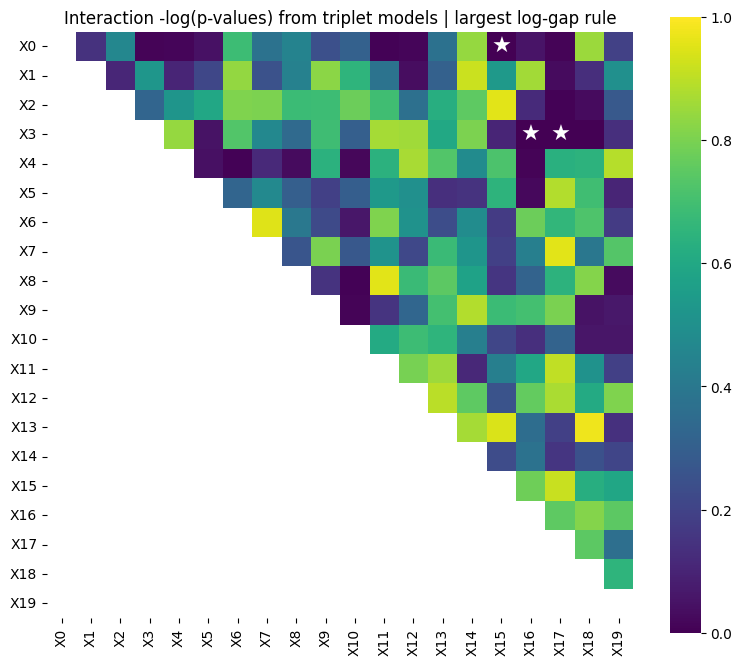
/Users/aymen20/Desktop/Research/repo/pkgs/uniPairs/.venv/lib/python3.9/site-packages/adelie/matrix.py:648: UserWarning: Detected matrix to be C-contiguous. Performance may improve with F-contiguous matrix.
warnings.warn(
100%|██████████| 102/102 [00:00:00<00:00:00, 32191.05it/s] [dev:6.2%]
100%|██████████| 100/100 [00:00:00<00:00:00, 35373.19it/s] [dev:4.7%]
Constructing interaction matrix with # pairs = 3
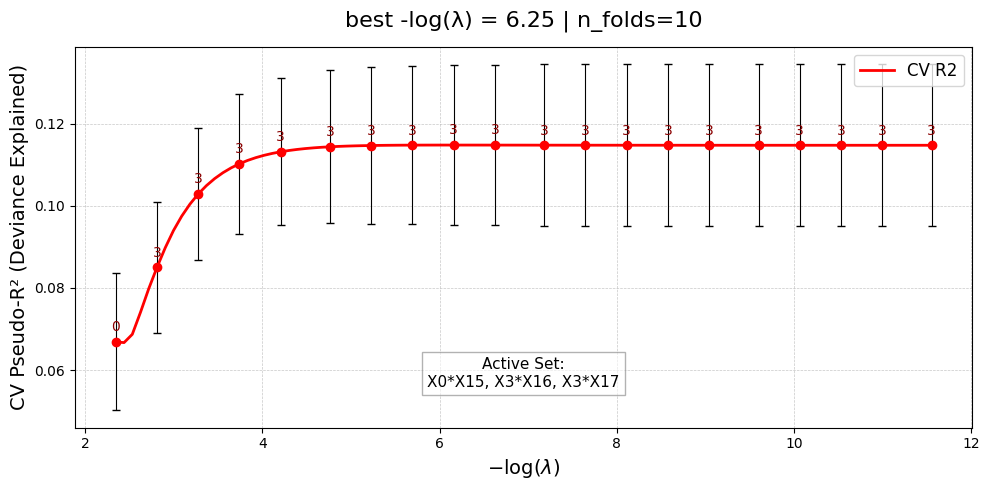
[Stage 2] Done. Active interactions: 3/3
=== UniPairs-2stage fit complete ===
Constructing interaction matrix with # pairs = 3
Constructing interaction matrix with # pairs = 3
UniPairsTwoStage/Test_ROC_AUC: 0.6732929400215645
UniPairsTwoStage/Train_ROC_AUC: 0.7614357101672111
UniPairsTwoStage/Coverage: 0.6666666666666666
UniPairsTwoStage/Model_size: 16
UniPairsTwoStage/FDP: 0.5
UniPairsTwoStage/Formula: -0.087 + 0.108*X0 + 0.321*X1 + 0.259*X2 + 0.126*X3 + 0.187*X4 + 0.074*X5 + -0.068*X7 + -0.016*X8 + -0.122*X10 + -0.118*X14 + -0.008*X15 + -0.008*X16 + -0.010*X17 + -0.052 + 0.367*X0*X15 + 0.190*X3*X16 + 0.236*X3*X17
# -----------------------------
# Fitting UniPairs
# -----------------------------
model = UniPairs(
two_stage=False,
lmda_path=None,
n_folds=10,
plot_cv_curve=True,
cv1se=False,
verbose=True,
interaction_candidates=None,
interaction_pairs=None,
family_spec={'family':'binomial'}
)
model.fit(X_train, Y_train)
pred_active_vars = model.get_active_variables()
formula = model.get_fitted_function()
alg_results = {
'UniPairsOneStage/Test_ROC_AUC': roc_auc_score(Y_test, model.predict(X_test, response_scale=True)),
'UniPairsOneStage/Train_ROC_AUC': roc_auc_score(Y_train, model.predict(X_train, response_scale=True)),
'UniPairsOneStage/Coverage': len(set(pred_active_vars) & set(true_active_vars)) / len(true_active_vars),
'UniPairsOneStage/Model_size': len(pred_active_vars),
'UniPairsOneStage/FDP' : len(set(pred_active_vars) - set(true_active_vars)) / len(pred_active_vars) if len(pred_active_vars)>0 else 0,
'UniPairsOneStage/Formula' : formula,
}
for k, v in alg_results.items():
print(f"{k}: {v}")
=== Starting UniPairs fit with 20 features ===
Fitting triplet models ...
Fitting 190 triplet models...
Progress: 19/190 triplets fitted...
Progress: 38/190 triplets fitted...
Progress: 57/190 triplets fitted...
Progress: 76/190 triplets fitted...
Progress: 95/190 triplets fitted...
Progress: 114/190 triplets fitted...
Progress: 133/190 triplets fitted...
Progress: 152/190 triplets fitted...
Progress: 171/190 triplets fitted...
Progress: 190/190 triplets fitted...
Triplet models complete. 0 unstable.
Scanning interactions ...
Selected 3 interaction pairs. (largest log-gap rule)


/Users/aymen20/Desktop/Research/repo/pkgs/uniPairs/.venv/lib/python3.9/site-packages/adelie/matrix.py:648: UserWarning: Detected matrix to be C-contiguous. Performance may improve with F-contiguous matrix.
warnings.warn(
100%|██████████| 100/100 [00:00:00<00:00:00, 4798.26it/s] [dev:62.0%]
100%|██████████| 101/101 [00:00:00<00:00:00, 7400.92it/s] [dev:52.9%]
Constructing interaction matrix with # pairs = 3
Cross-validating UniLasso ...
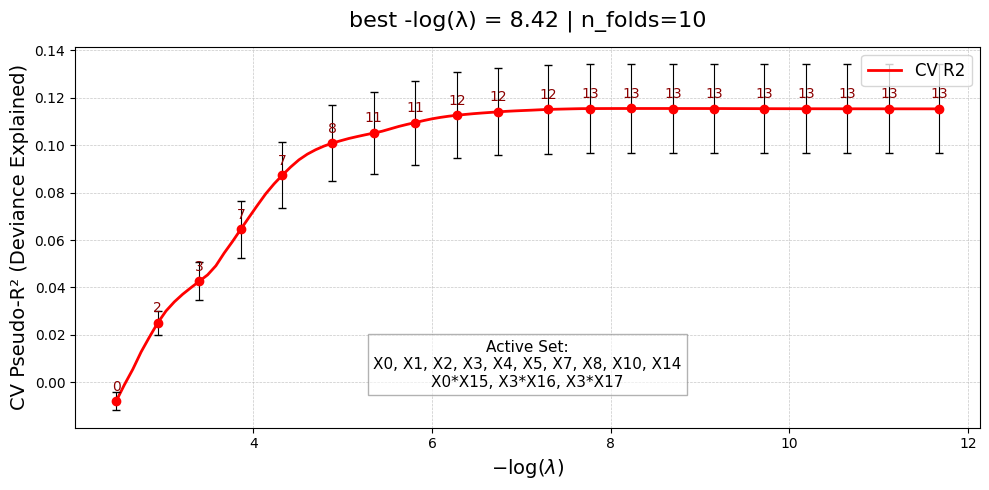
=== UniPairs fit complete ===
Constructing interaction matrix with # pairs = 3
Constructing interaction matrix with # pairs = 3
UniPairsOneStage/Test_ROC_AUC: 0.6710962519512707
UniPairsOneStage/Train_ROC_AUC: 0.7621083990005766
UniPairsOneStage/Coverage: 0.6666666666666666
UniPairsOneStage/Model_size: 16
UniPairsOneStage/FDP: 0.5
UniPairsOneStage/Formula: -0.089 + 0.135*X0 + 0.318*X1 + 0.311*X2 + 0.140*X3 + 0.230*X4 + 0.084*X5 + -0.102*X7 + -0.005*X8 + -0.108*X10 + -0.171*X14 + -0.009*X15 + -0.009*X16 + -0.010*X17 + 0.421*X0*X15 + 0.229*X3*X16 + 0.248*X3*X17